TRIBES: Relatedness detection in genomic data
We’ve developed TRIBES, a user-friendly platform for relatedness detection in genomic data.
The challenge
When genomics researchers conduct studies on large groups of people, people in the samples are often related. But sometimes they don’t even know it.
Knowledge about the relatedness of people in a sample is essential for research, particularly for disease gene discovery and genome wide association studies.
However, no current relatedness tools are accurate beyond 3rd degree (e.g., first cousins) and combine the necessary data processing steps for accuracy and ease of use.
Our response
We collaborated with Macquarie University to develop TRIBES, a user-friendly end-to-end package able to accurately detect up to 7th degree relatives (i.e., second cousins once removed).
Its novel approach for masking artefactual IBD segments also makes it the first tool to natively support disease gene mapping using ancestry information.
The platform includes built-in data pruning and phasing that previously would need to be performed using multiple tools. TRIBES is multithreaded and through the use of Snakemake is reproducible, customisable and executable on different compute environments (server, cluster and cloud).
Benefits
TRIBES is accurate past 3rd degree. In fact, it shows 99% accuracy (allowing 1 degree of error) for up to 7th degree relatives. This is a major improvement on commonly used tools like KING and PLINK, which are only accurate up to 3rd degree.
The platform is user-friendly and flexible, enabling:
- pruning
- phasing of genomes
- IBD segment recovery
- masking of artefactual IBD segments
- relationship estimation.
TRIBES facilitates disease locus discovery by offering a masking function to exclude artefactual IBD regions. By masking these regions, TRIBES both improves the accuracy of relationship estimates and is capable of highlighting a disease locus.
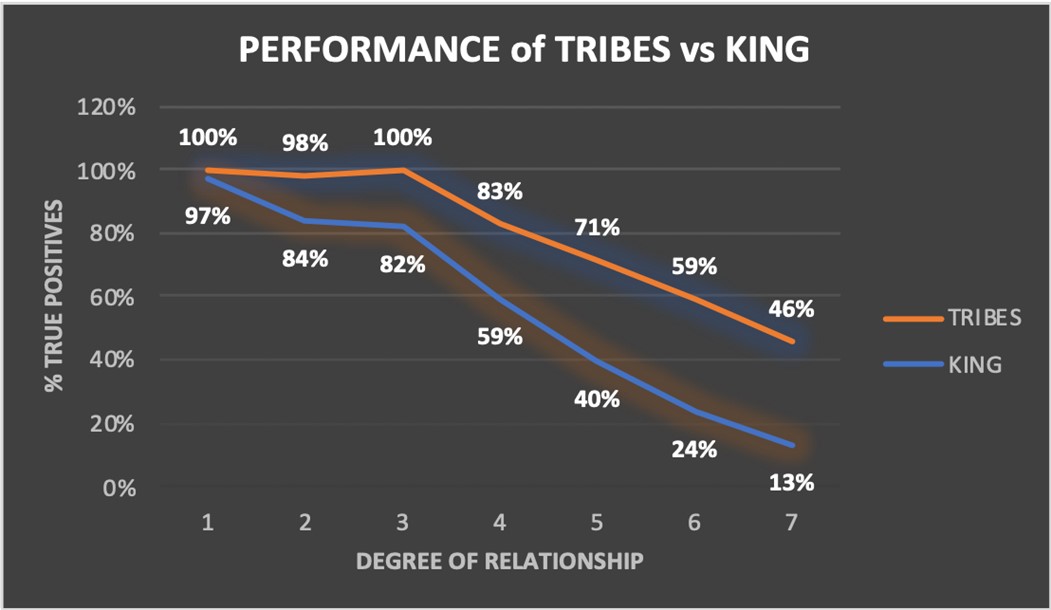
TRIBES outperforms gold standard tool KING at each degree of relationship, in terms of percentage of true positives detected in an ALS dataset with known relationships
The Australian e-Health Research Centre (AEHRC) is CSIRO's digital health research program and a joint venture between CSIRO and the Queensland Government. The AEHRC works with state and federal health agencies, clinical research groups and health businesses around Australia.

