GenPhenlnsights – Joining genomic and phenotypic data on-the-fly
“Going beyond research-scale solutions to realise the promise of genomics-driven precision medicine”
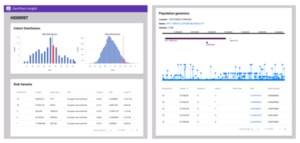
An example of the data produced by GenPhenInsight
The Challenge:
Stratifying patient cohorts by medical phenotype information, such as disease progression, has become a common practice to better understand treatment responses. However, doing the same on genomic data, which could give information on drug efficacy or risk to develop adverse reactions, is a substantial challenge because of the data sizes involved. Genomic data can be multiple gigabytes per subject and terabytes for larger cohorts. Even selectively aggregating genomic information over phenotypic sub-groups is currently not feasible to perform on the fly, which hampers clinical data exploration.
The Response:
In collaboration with QIMR Berghofer Medical Research Institute, the AEHRC has developed a cloud-native serverless framework that can seamlessly join and aggregate information over phenotypes and/or genotypes. The framework, called GenPhenInsight, was highlighted on the AWS Public Sector Blog to introduce a novel architecture able to handle both, compute and data intensive tasks. The system avoids data silos while preserving data ownership and patient privacy. It does this by maintaining a separation between patients’ medical information and their genomic information, as well as isolating data across separate S3 buckets, which allows institutions to tightly control information release.
Year Completed: 2018
The Australian e-Health Research Centre (AEHRC) is CSIRO's digital health research program and a joint venture between CSIRO and the Queensland Government. The AEHRC works with state and federal health agencies, clinical research groups and health businesses around Australia.
